Leading research organizations and computer manufacturers in the U.S. are collaborating on the construction of some of the world’s fastest supercomputers — exascale systems capable of performing more than a billion billion operations per second. A billion billion (also known as a quintillion or 1018) is about the number of neurons in ten million human brains.
The fastest supercomputers today solve problems at the petascale, meaning they can perform more than one quadrillion operations per second. In the most basic sense, exascale is 1,000 times faster and more powerful. Having these new machines will better enable scientists and engineers to answer difficult questions about the universe, advanced healthcare, national security and more.
At the same time that the hardware for the systems is coming together, so too are the applications and software that will run on them. Many of the researchers developing them — members of the U.S. Department of Energy’s (DOE) Exascale Computing Project (ECP) — recently published a paper highlighting their progress so far.
“The environment will really allow individual researchers to scale up their use of DOE supercomputers on deep learning in a way that’s never been done before.” — Rick Stevens, Argonne associate laboratory director for Computing, Environment and Life Sciences
DOE’s Argonne National Laboratory, future home to the Aurora exascale system, is a key partner in the ECP; its researchers are involved in not only developing applications, but also co-designing the software needed to enable applications to run efficiently.
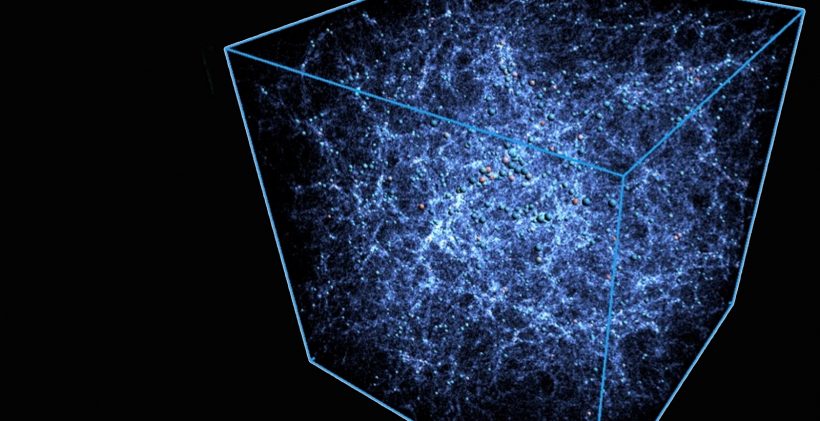
Computing the sky at extreme scales
One exciting application is the development of code to efficiently simulate “virtual universes” on demand and at high fidelities. Cosmologists can use such code to investigate how the universe evolved from its early beginnings.
High-fidelity simulations are particularly in demand because more large-area surveys of the sky are being done at multiple wavelengths, introducing more and more layers of data that existing high-performance computing (HPC) systems can’t predict in sufficient detail.
Through an ECP project known as ExaSky, researchers are extending the abilities of two existing cosmological simulation codes: HACC and Nyx.
“We chose HACC and Nyx deliberately because they have two different ways of running the same problem,” said Salman Habib, director of Argonne’s Computational Science division. “When you are solving a complex problem, things can go wrong. In those cases, if you only have one code, it will be hard to see what went wrong. That’s why you need another code to compare results with.”
To take advantage of exascale resources, researchers are also adding capabilities within their codes that didn’t exist before. Until now, they had to exclude some of the physics involved in the formation of the detailed structures in the universe. But now they have the opportunity to do larger and more complex simulations that incorporate more scientific input.

“Because these new machines are more powerful, we’re able to include atomic physics, gas dynamics and astrophysical effects in our simulations, making them significantly more realistic,” Habib said.
To date, collaborators in ExaSky have successfully incorporated gas physics within their codes and have added advanced software technology to analyze simulation data. Next steps for the team are to continue adding more physics, and once ready, test their software on next-generation systems.
Online data analysis and reduction
At the same time applications like ExaSky are being developed, researchers are also co-designing the software needed to efficiently manage the data they create. Today, HPC applications already output huge amounts of data, far too much to efficiently store and analyze in its raw form. Therefore, data needs to be reduced or compressed in some manner. The process of storing data long term, even after it is reduced or compressed, is also slow compared to computing speeds.
“Historically when you’d run a simulation, you’d write the data out to storage, then someone would write the code that would read the data out and do the analysis,” said Ian Foster, director of Argonne’s Data Science and Learning division. “Doing it step-by-step would be very slow on exascale systems. Simulation would be slow because you’re spending all your time writing data in and analysis would be slow because you’re spending your time reading all the data back in.”
One solution to this is to analyze data at the same time simulations are running, a process known as online data analysis or in situ analysis.
An ECP center known as the Co-Design Center for Online Data Analysis and Reduction (CODAR) is developing both online data analysis methods, as well as data reduction and compression techniques for exascale applications. Their methods will enable simulation and analysis to happen more efficiently.
CODAR works closely with a variety of application teams to develop data compression methods, which store the same information but use less space, and reduction methods, which remove data that is not relevant.
“The question of what’s important varies a great deal from one application to another, which is why we work closely with the application teams to identify what’s important and what’s not,” Foster said. “It’s OK to lose information, but it needs to be very well controlled.”
Among the solutions the CODAR team has developed is Cheetah, a system that enables researchers to compare their co-design approaches. Another is Z-checker, a system that lets users evaluate the quality of a compression method from multiple perspectives.
Deep learning and precision medicine for cancer treatment
Exascale computing also has important applications in healthcare, and the DOE, National Cancer Institute (NCI) and the National Institutes of Health (NIH) are taking advantage of it to understand cancer and the key drivers impacting outcomes. To do this, the Exascale Deep Learning Enabled Precision Medicine for Cancer project is developing a framework called CANDLE (CANcer Distributed Learning Environment) to address key research challenges in cancer and other critical healthcare areas.
CANDLE is a code that uses a kind of machine learning algorithm known as neural networks to find patterns in large datasets. CANDLE is being developed for three pilot projects geared toward (1) understanding key protein interactions, (2) predicting drug response and (3) automating the extraction of patient information to inform treatment strategies.
Each of these problems is at different scale — molecular, patient and population levels — but all are supported by the same scalable deep learning environment in CANDLE. The CANDLE software suite broadly consists of three components: a collection of deep neural networks that capture and represent the three problems, a library of code adapted for exascale-level computing and a component that orchestrates how work will be distributed across the computing system.
“The environment will really allow individual researchers to scale up their use of DOE supercomputers on deep learning in a way that’s never been done before,” said Rick Stevens, Argonne associate laboratory director for Computing, Environment and Life Sciences.
Applications such as these are just the tipping point. Once these systems come online, the potential for new capabilities will be endless.
Laboratory partners involved in ExaSky include Argonne, Los Alamos and Lawrence Berkeley National Laboratories. Collaborators working on CANDLE include Argonne, Lawrence Livermore, Los Alamos and Oak Ridge National Laboratories, NCI and the NIH
The paper, titled “Exascale applications: skin in the game” is published in Philosophical Transactions of the Royal Society A.
CANDLE was first described in the following publication. Since then, many significant improvements and new functionality have been added. Wozniak, Justin M., Rajeev Jain, Prasanna Balaprakash, Jonathan Ozik, Nicholson T. Collier, John Bauer, Fangfang Xia, Thomas Brettin, Rick Stevens, Jamaludin Mohd-Yusof, Cristina Garcia Cardona, Brian Van Essen, and Matthew Baughman 2018. “CANDLE/Supervisor: A Workflow Framework for Machine Learning Applied to Cancer Research.” BMC Bioinformatics 19 (18): 491. https://doi.org/10.1186/s12859-018-2508-4.
The Exascale Computing Project is part of the DOE-led Exascale Computing Initiative (ECI). ECI is a partnership between DOE’s Office of Science and the National Nuclear Security Administration.
Argonne National Laboratory seeks solutions to pressing national problems in science and technology. The nation’s first national laboratory, Argonne conducts leading-edge basic and applied scientific research in virtually every scientific discipline. Argonne researchers work closely with researchers from hundreds of companies, universities, and federal, state and municipal agencies to help them solve their specific problems, advance America’s scientific leadership and prepare the nation for a better future. With employees from more than 60 nations, Argonne is managed by UChicago Argonne, LLC for the U.S. Department of Energy’s Office of Science.
The U.S. Department of Energy’s Office of Science is the single largest supporter of basic research in the physical sciences in the United States and is working to address some of the most pressing challenges of our time. For more information, visit https://energy.gov/science.